Beyond IC50: Binding kinetics in drug discovery
Drugs with similar affinity can exhibit very different kinetic and binding mechanism profiles, which may contribute to efficacy, safety, duration of action and differentiation from other similar therapies.
Early evaluation of the kinetics of drug-target interactions helps to improve decision-making, to focus on the most promising compounds.
Binding kinetics define efficacy of target occupancy
- Onset of the drug action is influenced by the association rate (kon): the faster a drug binds to the target, the faster the occupation will occur.
- Duration of drug action is dependent on the dissociation rate (koff): the longer the duration of binding, the longer the pharmacodynamic effect.
- Differences in association and dissociation rates are not identified by affinity measurement alone.
Applications
Predict drug clinical efficacy: By using binding kinetics, we help our clients find the most effective compounds early.
In biochemical and cellular assays performed at equilibrium, the drug and target are present at constant concentrations and thus the IC50 accurately reflects the concentration of the drug-target complex. However, in the body the concentration of a drug constantly changes due to several physiological processes. Hence, affinity is not the only relevant parameter that predicts target occupancy and ultimately efficacy: clear understanding of binding kinetics provides a more complete picture of what drives target occupancy in
vivo1-6.
The correlation of in vivo efficacy with residence time has been demonstrated across a spectrum of target types.
| Target class | Target | References |
| GPCRs | A3R | Xia L. et al. (2017) J Med Chem. 14;60(17):7555-7568. |
| GPCRs | A2AR | Hothersall J.D. et al. (2017) Mol Pharmacol. 91(1):25-38. |
| GPCRs | β2AR | Rosethorne E.M. et al (2016) Mol Pharmacol. 89(4):467-75. |
| GPCRs | CB1 | Louvel J. et al (2015) Eur J Med Chem. 28;101:681-91. |
| GPCRs | CCR2 | Vilums M. et al (2015) ChemMedChem. 10(7):1249-58. |
| GPCRs | CCR5 | Swinney D.C. et al. (2014) Br J Pharmacol. 171(14):3364-75. |
| GPCRs | HRH1 | Bosma R. et al. (2017) Front Pharmacol. 25;8:667. |
| GPCRs | GIPR | Nordskov M.B.G et al. (2020) Basic Clin Pharmacol Toxicol. 126 Suppl 6:122-132. |
| GPCRs | NK1R | Nederpelt I. et al. (2017) Sci Rep. 26;7(1):14169. |
| GPCRs | mGluR2 | Doornbos M.L.J. et al. (2017) J Med Chem. 10;60(15):6704-6720. |
| GPCRs | M3 mAChR | Guo D. et al. (2018) Methods Mol Biol. 1705:197-206. |
| Nuclear receptors | ER | Copeland R.A. (2016) Nat Rev Drug Discov. 15(2):87–95. |
| Kinases | ABL | Puttini M. et al. (2008) Haematologica 93, 653–661. |
| Kinases | AURK B, AURK C | Anderson K. et al. (2009) Biochem J. 420(2):259-65. |
| Kinases | BTK | Bradshaw J.M. et al. (2015) Nat Chem Biol. 11:525–531. |
| Kinases | CDK2, CDK9 | Ayaz P. et al. (2016) ACS Chem Biol 11(6):1710-9. |
| Kinases | CDK7 | Marineau J.J. et al. (2021). doi: 10.1021/acs.jmedchem.1c01171. |
| Kinases | EGFR | Wood E.R. et al. (2004) Cancer Res. 64, 6652–6659. |
| Kinases | p38α MAPK | Walter N.M. et al. (2017) J Med Chem. 12;60(19):8027-8054. |
| Kinases | TTK | Uitdehaag J.C.M. et al. (2017) J Mol Biol. 7;429(14):2211-2230. |
| Kinases | RIP1K | Yoshikawa M. et al (2018) J Med Chem. 61:2384–2409. |
| Proteases | AChE | Kharlamova A.D. et al (2016) Biochem J. 473(9):1225-36. |
| Proteases | BACE1 | Eketjäll S. et al. (2016) J Alzheimers Dis. 50(4): 1109–1123. |
| Epigenetic enzymes | DOT1L | Daigle S.R. et al (2013) Blood. 122(6): 1017–1025. |
| Epigenetic enzymes | HDAC1, HDAC2 | Wagner F.F. et al. (2016) Bioorg Med Chem. 15;24(18):4008-4015. |
| Epigenetic enzymes | EZH2 | Van Aller G.S. et al. (2014) ACS Chem Biol. 21;9(3):622-9. |
| Synthases | COX-2 | Tian G. et al. (2021) Biochemistry 60(31):2407-2418. |
| Ion channels | L-type calcium | Copeland R.A. (2016) Nat Rev Drug Discov. 15(2):87–95. |
| Ion channels | M2 proton | Drakopoulos A. et al. (2018) ACS Med Chem Lett. 9(3):198-203. |
Comprehensive reviews of the importance of drug-target kinetics
- Copeland R.A. (2016) Nat Rev Drug Discov. 15(2):87–95.
- Doris A Schuetz D.A. (2017) Drug Discov Today. 22(6):896-911.
- Swinney D.C. (2004) Nat Rev Drug Discov. 3(9):27–41.
- Zhang R. et al. (2010) Expert Opin Drug Discov. 5(11):1023-9.
- Tummino P.J. et al. (2008) Biochemistry. 47(20):5481–5492.
- Lu H. at al. (2010) Curr Opin Chem Biol. 14(4):467–474.
Extend the duration of action: The longer a drug remains bound to its target, the longer it remains active, meaning less frequent doses are needed.
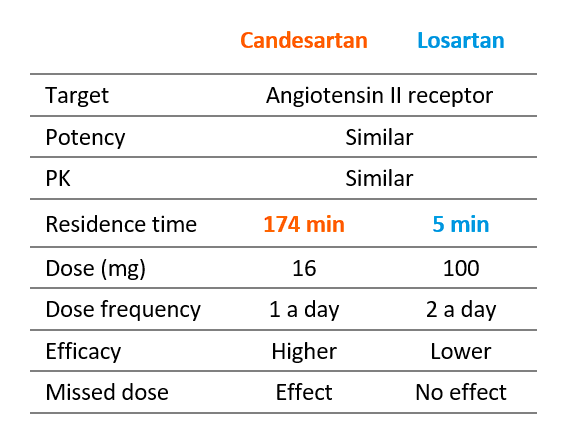
There are many medicines for which binding kinetics contribute to efficacy and duration of action. One example is Candesartan, an antihypertensive drug that lowers blood pressure more efficiently than any other member of the same class, such as Losartan.
While Candesartan and Losartan share the same binding site, similar pharmacokinetics and receptor occupancy profiles, their binding kinetics are completely different1-4. In a clinical trial study, the long-lasting pharmacodynamic effect of Candesartan provided greater efficacy at a lower dose (16 mg) than Losartan (100 mg) and maintained efficacy in the event of a missed dose5.
Left: Dose–response curves for the effect of Candesartan (orange) and Losartan (blue) on blood pressure in hypertensive patients. Right: Pharmacodynamic, pharmacokinetic and clinical features of Candesartan and Losartan. Adapted from Elmfeldt D. et al (2002).
References
- Vauquelin G. et al (2010) Br J Pharmacol. 161(3): 488–508.
- Tummino P.J. et al. (2008) Biochemistry. 47(20):5481–5492.
- Copeland R.A. et al. (2006) Nat Rev Drug Discov 5(9):730-739.
- Swinney D.C. (2004) Nat Rev Drug Discov. 3(9):27–41.
- Elmfeldt D. et al (2002) Blood Press. 11(5):293-301.
Compound differentiation: Kinetic profiling in the early stages of drug discovery enables better identification of compounds with therapeutic potential that merit further progression.
Simple affinity measurements may mask the true impact of structural changes made in the same chemical series.
Ayaz and colleagues show this with a series of Roniciclib analogs of CDK21. They found that Roniciclib, the trifluoromethyl derivatives (CF3) and bromo analogues (Br) showed similar kinase and cell proliferation inhibition along with comparable pharmacokinetic profiles. However, Roniciclib and the CF3 derivatives had longer residence times and superior efficacy in tumor growth inhibition relative to the corresponding bromo analogues.
Left: Comparison of the residence times of compounds 1–5 and Roniciclib. The CF3 derivatives (orange) have up to 15 times longer residence times than their Br analogues (blue). Right: Comparison of cell proliferation and antitumor efficacy. The CF3-substituted compounds show superior efficacy in tumor growth inhibition relative to the Br analogs despite their similar EC50 values. Adapted from Ayaz, P. et al. (2016).
References
- Ayaz, P. et al. (2016) ACS Chem. Biol. 11, 1710–1719.
Maximize therapeutic windows: Early modulation of on- and off-target kinetics can help build safety margins and reduce adverse events.
It is now understood that drug safety is difficult to achieve by solely considering affinity. Indeed, many reports link selectivity to the kinetic profile of the drugs.
Find out more by reading the appropriate case study:
Affinity doesn’t tell the whole story of MET inhibitor Foretinib
Understand PK/PD disconnects: Kinetic profiling often bridges the gap between PK/PD prediction and observation.
Systemic exposure as a driver of pharmacologic effects provides limited value in guiding compound optimization, selection, and advancement when PK/PD disconnects are observed within a chemical series. Binding kinetics can help in this scenario not only guiding compound design but also predicting target occupancy and drug dose.
Build better models: PK/PD models that integrate kinetics better define therapeutic windows and therefore dose regimens.
Client feedback and several studies1-8 suggest that the pharmacokinetic/pharmacodynamic models that integrate binding kinetics, instead of just affinity, better predict:
- Target engagement.
- Drug dose.
- Treatment schedule.
- Real selectivity and potential toxicities.
Simulations of the time course of in vivo target occupancy (TO) by Dasatinib show that conventional PK/PD approaches predict lower efficacy and a higher dose than is actually needed. Affinity based models also fail to predict the real selectivity and consequently, potential toxicities.

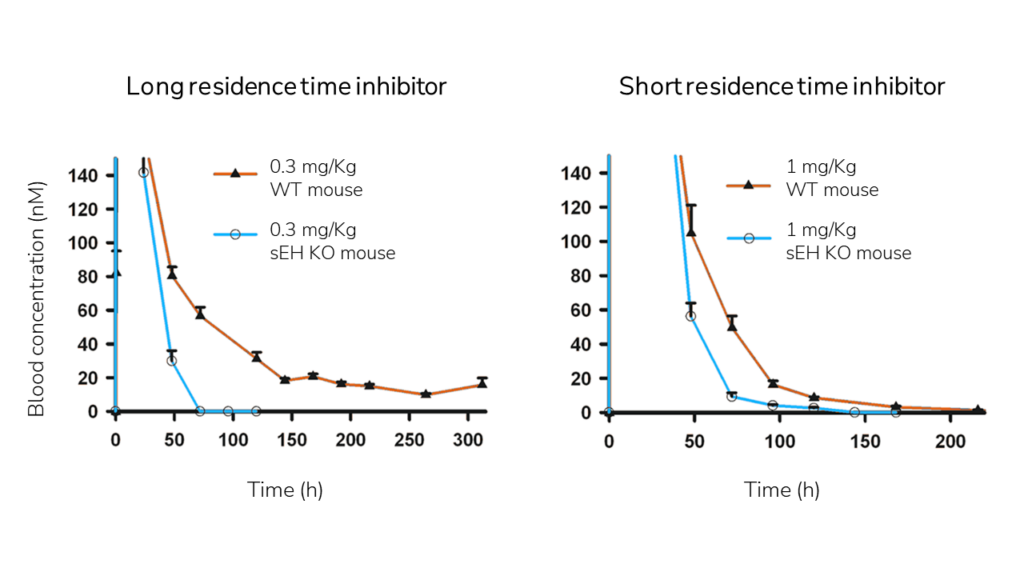
This effect of binding kinetics on drug PK has been demonstrated in mice. The comparison of the PK profile of inhibitors with similar Kd but different residence time between the WT and epoxide hydrolase (EH) knockout mice, reveals that drugs with longer residence time bind to EH longer, protecting them from being metabolized or eliminated from the body.
References
- Georgi V. et al (2018) J Am Chem Soc. 140(46):15774–15782.
- Daryaee F. et al (2017) Chem Sci. 8(5): 3434–3443.
- Walkup G.K. et.al (2015) Nat Chem Biol. 11(6):416–423.
- Lee K.S.S. et al. (2019) ACS Central Sci. 5(9):1614–1624.
- De Witte W.E.A. (2018) J Pharmacokinet Pharmacodyn. 45(4):621-635.
- Ramsey J.S. et al (2011) Br J Pharmacol. 164(3):992-1007.
- Hong Y. et al. (2008) Clin Pharmacokinet. 47(2):129-37.
- Yassen A. et al (2005) J Pharmacol Exp Ther. 313(3):1136-49.
Strengthen intellectual property: By using binding kinetics, our clients enrich the structural diversity of their pipeline
Understanding binding kinetics early in drug discovery gives access to a more diverse chemical space, better defined biology and therefore more scope for intellectual property.
Technology
KINETICfinder®
KINETICfinder® is Enzymlogic’s patented high-throughput screening platform that delivers all key binding kinetic (kon / koff / residence time) and affinity (kd) parameters for reversible binders quickly and at scale.
Our highly robust platform provides accurate and reproducible compound–target interaction data that enables improved screening of compounds in early discovery: to iterate medicinal chemistry, understand PK/PD disconnects and build better models to define therapeutic windows.
List of targets ready to use
Our expertise extends beyond kinases to GPCR and other target types.
Search our database to discover whether your target is:- May be possible: We have a strong track record of achievement where others fail! Contact us to find out more.
- Clear line of sight: Establishment requires finalization / validation, which typically takes 2-4 weeks to complete.
- Good to go: Data within 2 weeks of compound receipt.
* Full length and partial length forms are available.
| Target name | Alternative names | KINETICfinder | COVALfinder |
|---|---|---|---|
| AAK1 | KIAA1048, DKFZp686K16132 | Clear line of sight | |
| ABL1 | ABL, c-ABL1, BCR-ABL, CHDSKM, JTK7, bcr/abl, c-ABL, p150, v-abl | Good to go | |
| ABL1 E255K | ABL, c-ABL1, BCR-ABL, CHDSKM, JTK7, bcr/abl, c-ABL, p150, v-abl | Clear line of sight | |
| ABL1 G250E | ABL, c-ABL1, BCR-ABL, CHDSKM, JTK7, bcr/abl, c-ABL, p150, v-abl | Clear line of sight | |
| ABL1 H396P | ABL, c-ABL1, BCR-ABL, CHDSKM, JTK7, bcr/abl, c-ABL, p150, v-abl | Clear line of sight | |
| ABL1 M351T | ABL, c-ABL1, BCR-ABL, CHDSKM, JTK7, bcr/abl, c-ABL, p150, v-abl | Clear line of sight | |
| ABL1 Q252H | ABL, c-ABL1, BCR-ABL, CHDSKM, JTK7, bcr/abl, c-ABL, p150, v-abl | Clear line of sight | |
| ABL1 Y253F | ABL, c-ABL1, BCR-ABL, CHDSKM, JTK7, bcr/abl, c-ABL, p150, v-abl | Clear line of sight | |
| ABL2 (ARG) | ARG, ABL isoform b, ABLL | Good to go | |
| ACK (TNK2) | TNK2, ACK-1, ACK1, p21cdc42Hs | Good to go | |
| ACVR2A | ACTRII, ACTR2, ACVR2 | Clear line of sight | |
| ACVR2B | ACTRIIB, ActR-IIB, HTX4 | Clear line of sight | |
| AKT1 | AKT1, PKB, PKB-ALPHA, PRKBA, RAC, RAC-ALPHA, | Good to go | |
| ALK | ALK, CD246, NBLST3 | Good to go | May be possible |
| ALK C1156Y | ALK, CD246, NBLST3 | Clear line of sight | |
| ALK F1174L | ALK, CD246, NBLST3 | Clear line of sight | |
| ALK L1196M | ALK, CD246, NBLST3 | Clear line of sight | |
| ALK R1275Q | ALK, CD246, NBLST3 | Clear line of sight | |
| ALK T1151_L1152insT | ALK, CD246, NBLST3 | Clear line of sight | |
| ALK1 (ACVRL1) | ALK1, ALK-1, ACVRLK1, HHT, HHT2, ORW2, SKR3, TSR-I | Good to go | May be possible |
Helpful resources

How can we help?
Our experienced project manager will work with you to identify the best way forward for your project.